|
Davis, California, February 10, 2004 -- Allometra announced the release of Arabidopsis FASTA files with annotation incorporating MPSS
expression data. The files are freely available for download from:
http://allometra.com/ath_fasta_mpss.shtml
 Arabidopsis MPSS data for five tissues: callus, flower, leaf, root and silique were incorporated into the annotation of Arabidopsis
genes. The data for unique detected signatures from the public Arabidopsis MPSS project (University of Delaware) were appended to the
annotation. The primary reason for producing these files was their easy utilization in Allometra's genomics data analysis program,
PyMood, for sophisticated querying of BLAST output data.
Arabidopsis MPSS data for five tissues: callus, flower, leaf, root and silique were incorporated into the annotation of Arabidopsis
genes. The data for unique detected signatures from the public Arabidopsis MPSS project (University of Delaware) were appended to the
annotation. The primary reason for producing these files was their easy utilization in Allometra's genomics data analysis program,
PyMood, for sophisticated querying of BLAST output data.
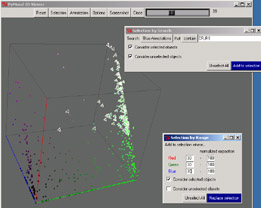
PyMood is a genomic data visualization program, which allows visual displaying of BLAST output files and expression
data. PyMood runs on Windows 2000/XP, Mac OS X, and Linux. The visualization principle in PyMood is coloring and
positioning objects (genes, proteins, etc.) in space according to their data values. The 3D output in PyMood can be
rotated, zoomed in and out, shifted, and queried.
The 2D Visualization capabilities of PyMood allow presentation of processed data file as a canvas comprised of the
colored elements, which represent individual genes or proteins. The color of the elements reflects their data values.
The canvas elements can be sorted in a two-dimensional plane according to their color (data) values. These visual
outputs can be automatically published: the program generates web pages with canvas and 2D plot images where every
element is clickable and linked to a database of the user's choice.
The PyMood BLAST Launcher allows running BLAST programs in a user-friendly environment; post-processing of BLAST outputs
(BLAST parser is built into the PyMood BLAST Launcher); automated creation of browsable web tables from parsed BLAST
data outputs, manipulations with FastA files: combining, splitting files and sequences, creation of non-redundant sets,
and more.
Additional information can be obtained by sending a request to: info@allometra.com
| |
|


