
 |

| home | contact | press | arabidopsis | yersinia | fowlpox | mpss | cacao |
Arabidopsis MPSS data for five tissues: callus, flower, leaf, root and silique were incorporated into the annotation of Arabidopsis genes. The MPSS data for unique detected signatures from the public Arabidopsis MPSS project (University of Delaware, Blake Meyers) were appended to the annotation. An example of the resulting annotation is displayed here: 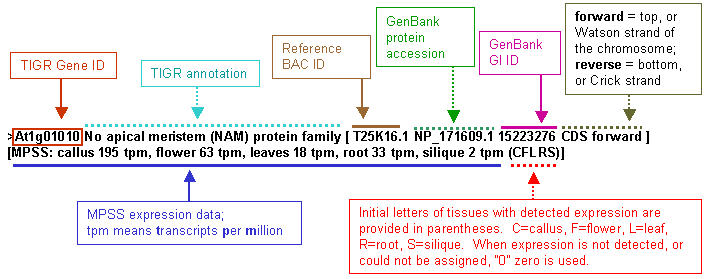 The primary reason for producing these files was their easy utilization in our genomics data analysis program, PyMood ( http://allometra.com ) , for sophisticated querying of BLAST output data. When utilizing this file in PyMood as a query or one of target datasets it is easy to query for genes that have expressed or not expressed homologs in Arabidopsis. For example, a search in PyMood through BLAST output data (when any of these files are used as one of the target databases) with a query “C0000” will retrieve the genes that have Arabidopsis homologs expressed in callus only. The files (in zip format): x-ath-ncbi-mpss-unspl.fasta [24MB] containing unspliced genes with 500 nucleotides preceding start codon and 500 nucleotides following stop codon. Exons are shown in capital letters. x-ath-ncbi-mpss-prot.fasta [7.4MB] contains translated to protein spliced exons. x-ath-ncbi-mpss-cds.fasta [11MB] contains all extracted CDS from start codon to stop codon without introns. Please note that these files contain TIGR Arabidopsis annotation version 4 (not the latest one), and include information about one (longest) splice variant for every gene only. More detailed MPSS data for every gene can be extracted from http://mpss.udel.edu/at/ | ||||||||||||||||||
|
|