Visualization of the Arabidopsis MPSS data for five tissues: callus, flower, leaf, root, and silique
|
|
Fasta MPSS Example
This is just one example of how these MPSS expression annotated FASTA files can be used in
PyMood.
- A novel cotton library (638 contigs) was compared using PyMood BLAST Launcher with:
- TIGR maize EST assembly (the red target database)
- All GenBank cotton nucleotide sequences (the green database)
- Arabidopsis proteins (the blue database)
- The cotton novel library was displayed in the PyMood 3D Viewer, so that every query gene is colored and positioned in space according to its level of homology to the best hits in the three target databases. The output is presented in Figure 1, where the selected elements are the genes from the cotton library that have conservative homologs (e< -30) in all three target databases, and at the same time, their Arabidopsis homologs are expressed in all five tissues.
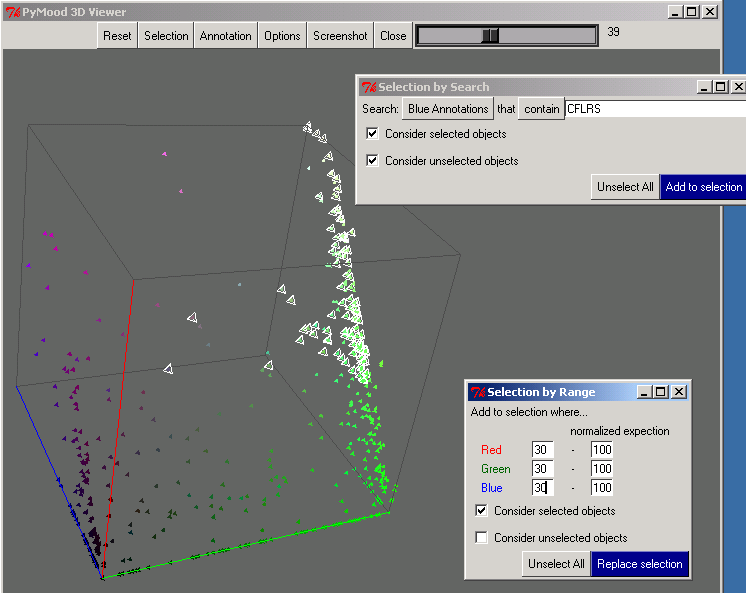 Figure 2 shows the PyMood 3D Annotation window where the annotation is displayed for every selected gene. The query gene annotation is displayed in black font, and the hits from the target databases are displayed in the font color assigned to the databases.
Figure 2 shows the PyMood 3D Annotation window where the annotation is displayed for every selected gene. The query gene annotation is displayed in black font, and the hits from the target databases are displayed in the font color assigned to the databases.
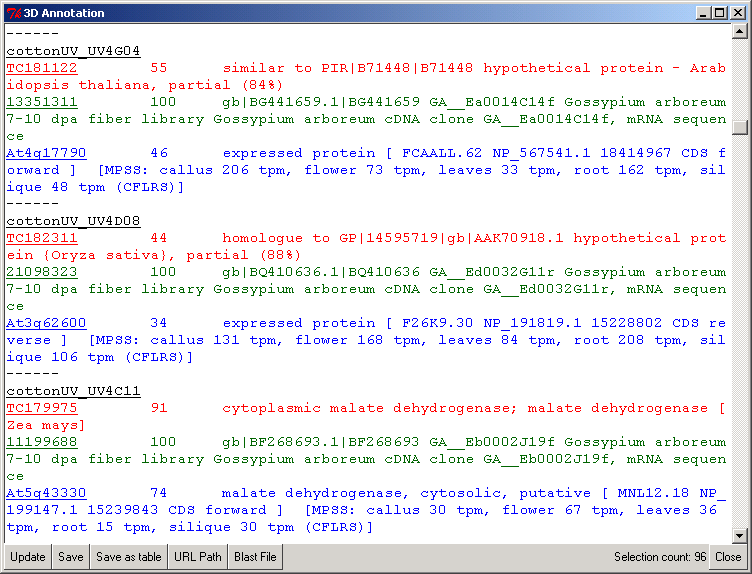 Table 1 (a tab delimited text file, can be opened in MS Excel) contains the information about all the 96 selected elements and their homologs.
Table 1 (a tab delimited text file, can be opened in MS Excel) contains the information about all the 96 selected elements and their homologs.
|




