Comparison of Yersinia pestis with Escherichia coli, Salmonella typhi, and Salmonella typhimurium
 |
|
|
|
(1.7MB) |
|
 |
gene lists
Text files extracted from 2D Plots
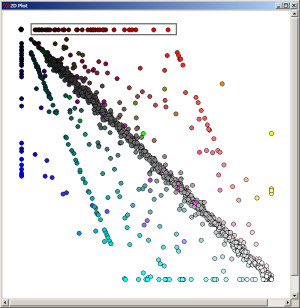
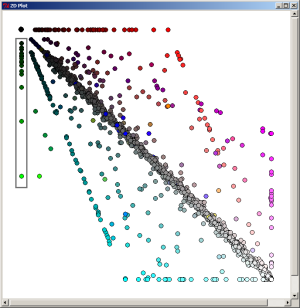
When the most-dark element is selected on the 2D Plot, the 1070 proteins which are specific (in the limit of our expectation cutoff in the PyMood BLAST Launcher, e-8) to Yersinia pestis are extracted.
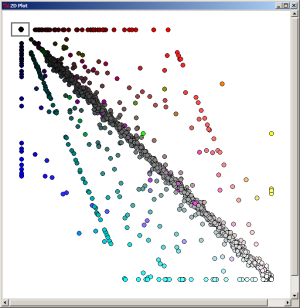
The list of the selected genes were saved as a text file:
Ypestis_specific.txt
|
By selecting the axially positioned elements, only proteins shared by the query database and one target database were retrieved. When the red axis elements are selected, the list of 93 proteins common to Y. pestis and E. coli were retreived.
Use the following link to download the list of the genes common to Yersinia pestis and E. coli only:
Ypestis_Ecoli_specific.txt
|
By selecting the axially positioned elements, only proteins shared by the query database and one target database were retrieved. When the green axis elements are selected, the list of 26 proteins common to Y. pestis and S. typhi were retreived.
Use the following link to download the list of the genes common to Yersinia pestis and S. typhi only:
Ypestis_Styphi_specific.txt
|
By selecting the axially positioned elements, only proteins shared by the query database and one target database were retrieved. When the blue axis elements are selected, the list of 52 proteins common to Y. pestis and S. typhimurium were retreived.
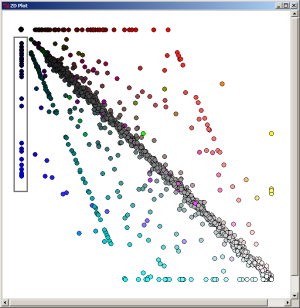
Use the following link to download the list of the genes common to Yersinia pestis and S. typhimurium only:
Ypestis_Styphimurium_specific.txt
|
Text files extracted from 3D Plots
When a group of the lightest elements (70 to 100 normalized expectation values) are selected on the 3D Plot, the most conservative proteins (within the analyzed group of organisms) are extracted from the data.
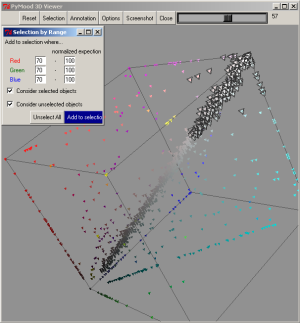
The list of the selected genes were saved as a text file:
1579.txt
|
When the elements with approximately equal values for all three coordinates are selected on the 3D Plot, the proteins evolving with about the same speed in Y. pestis, E. coli, S. typhi, and S. typhimurium are extracted. The selected elements are 2400 Y. pestis proteins which have equally distant homologs in three other species: E. coli, S. typhi, and S. typhimurium
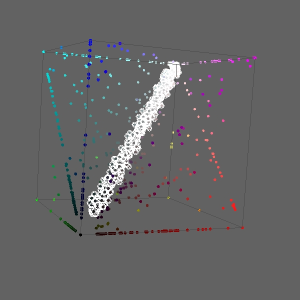
The list of the selected genes were saved as a text file:
2400.txt
|
The selected elements are 156 Y. pestis proteins which are absent in E. coli, but have equally distant homologs in S. typhi and S. typhimurium.
The list of the selected genes was saved as a text file:
156.txt
|
The selected elements are 49 Yersinia pestis proteins that present in E. coli, S. typhimurium, but absent in S. typhi.
The list of the selected genes were saved as a text file:
49.txt
|
The selected elements are 1256 proteins that are highly conservative between Yersinia pestis and E. coli, but has various but equal levels of similarity to the S. typhi and S. typhimurium homologs.
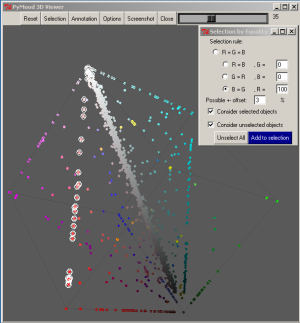
The list of the selected genes were saved as a text file:
1256.txt
|
The selected elements are 1260 proteins that are highly conservative between Yersinia pestis, S. typhi, and S. typhimurium, but have various levels of similarity to the corresponding E. coli homologs.
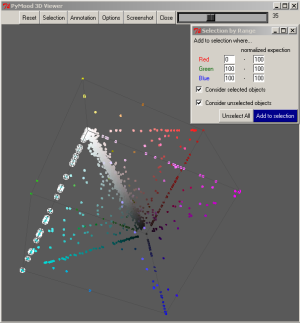
The list of the selected genes were saved as a text file:
1260.txt
|
In this search we retrieved 10 Yersinia pestis specific membrane proteins out of the 1070 Y. pestis species specific proteins. (When compared to E. coli, S. typhi, and S. typhimurium)
The list of the selected genes were saved as a text file:
10_Yspecific_membrane.txt
|
|


