
 |

| home | contact | press | arabidopsis | yersinia | fowlpox | mpss | cacao |
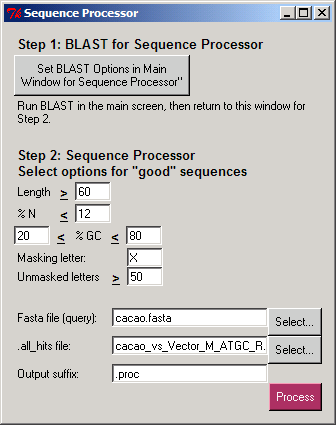 Davis, California, August 19, 2004 -- Allometra, LLC announces the release of PyMood 2 version 9. In this release the following new features are implemented:
Davis, California, August 19, 2004 -- Allometra, LLC announces the release of PyMood 2 version 9. In this release the following new features are implemented: 1. New function: Sequence Processor for sorting and masking of sequences in the PyMood BLAST Launcher. A detailed description of this function is available at: http://allometra.com/sequence_proc.shtml 2. New project: The "cocoa_allometra" project in the "projects" folder as a Sequence Processor sample. The info on this project is available at: http://allometra.com/cacao.shtml 3. Help Viewer: The old help viewer is removed and replaced with an html-based viewer. 4. Added checking for spaces in filenames in PyMood BLAST Launcher. 5. On Windows, NCBI BLAST is upgraded to version 2.2.9. 6. On Mac OS X Panther, colored axes are added to the 3D Viewer. PyMood is a plug-and-play desktop application for visualizing genomic data. It is used to analyze and display relations between complete genomes, genomic fragments, proteins, ESTs, full-length cDNAs, and virtually any sets of sequences. The program is also used to visualize and analyze expression data. The visualization principle in PyMood is coloring and positioning objects (genes, proteins, etc.) in space according to their data values. The 3D output in PyMood can be rotated, zoomed in and out, shifted, and queried. The produced outputs allow visual presentation of patterns in the database, so the user can form the queries based on observed patterns. The patterns and groups of genes can be easily retrieved using PyMood selection widgets. The program description and example projects are available at: http://allometra.com |
|
|